Projects
Single molecule fluorescence resonance energy transfer (smFRET) experiments exploit the distance dependency of the measurable transfer efficiency between two dyes to determine distances on the nm scale. The transfer efficiency in addition depends on the instantaneous mutual orientation inaccesible to experiments. In molecular dynamics simulations, the mutual orientation of the dyes is accessible. Therefore, combining simulations with experiments results in more accurate distances as estimated from experiments alone.
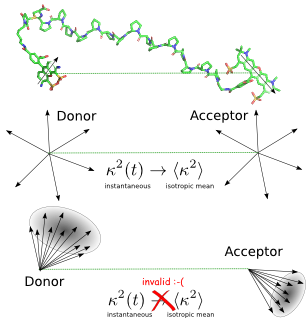
FRET Approximation
Single molecule fluorescence resonance energy transfer (smFRET) experiments measure the non-radiative transfer efficiency from a donor to an acceptor dye. The transfer efficiency depends on the inter-dye distance and is measured by monitoring the relative fluorescence intensities of donor and acceptor. Measurements of the FRET efficiency on individual molecules with dyes attached probe the distance in or between biomolecules and biomolecular subunits. The optical read-out of fluorescence allows application of the technique at ambient conditions and in vivo. The recent development of time-resolved (tr) experiments enables the extraction of information on single molecule dynamics. These advantages resulted in the emergence of smFRET as a powerful tool in biophysics providing a sensitive nanometer distance reporter to investigate biological structures and their function. Ligand-receptor interactions, conformational changes, protein synthesis and folding kinetics as well as translocation of genes are only some of the multitude of issues that can be addressed by tr-smFRET.
In analogy to television broadcasting, where the received signal strength depends on the antenna orientation, the transfer efficiency not only depends on the inter-dye distance, but also the mutual dye orientation. Therefore, the extraction of distance information from smFRET experiments is not straightforward and involves approximations. Typical approximations are are isotropic and uncorrelated dye transition dipole orientations and that the coupling potential is sufficiently described by dipole-dipole coupling of the transition densities. With these and further approximations, Theodor Förster expressed [1] the transfer efficiency as \[E(R) = \frac{1}{1+\left(\frac{R}{R_0}\right)^6} \qquad (1),\] where \(R\) is the inter-dye distance and \(R_0\) the distance with 50 % excitation transfer. The orientation dependency of dipole-dipole coupling is given by \[\kappa=\cos\theta_{DA}-3\cdot\cos\theta_D\cdot\cos\theta_A \qquad (2),\] with \(\theta_{DA}\) as angle between the two transition dipoles of the dyes. \(\theta_D\) and \(\theta_A\) are the angle between transition dipole of donor or acceptor and the dye separation vector \(R\), respectively. The Förster radius \(R_0^6\) is \(\propto \kappa^2\) and \(\langle\kappa^2\rangle=\frac{2}{3}\) for isotropic orientation distributions.
Dyes attached to biomolecules are sterically restricted and interact with the dye’s surface. In fact, strong deviations of the isotropicity and thus also of \(\langle\kappa^2\rangle\) from \(\frac{2}{3}\) were found [2]. Thus, accurate distance measurements with FRET are hindered by the uncertainty of the dye orientation. In contrast to experiments, the orientation of dyes is accessible at each instant in molecular dynamics simulations (MD).
Dye Dynamics and FRET Efficiency Distributions from MD simulations
Aiming at improving the accuracy of distance reconstruction, we developed a combination of dye orientation statistics and dynamics from MD simulations with experimentally measured efficiency distributions [3]. As test system, we studied poly-proline with two dyes attached at the ends. Poly-prolines form a stable type II helix in water and were extensively studied by FRET experiments in the past [4,5]. To mimic the kinetic pathways of smFRET experiments, we developed a Markov Chain Monte Carlo method with time dependent FRET rates calculated from MD trajectories.
Exemplary pathways in Markov Chain Monte Carlo. First the Donor is excited emitting a photon after a while. In the second case, the excitation is transfered to the acceptor dye via FRET and released as photon after a while as well.
Using the developed Markov Chain Monte Carlo for photon generation [6], photons bursts were generated as measured by the experiment. In analogy to the experiment, the donor and acceptor photon counts yield burst efficiencies and efficiency distributions can be compared with experimentally obtained ones. Simulations demonstrate, that the origin of the experimentally observed heterogeneity is twofold. First, poly-proline has a non-negligible isomerization probability and thus part of the species in the ensemble are cis-isomers with smaller end-to-end distance. Second, the dyes interact with the poly-proline backbone leading to open and closed conformations with different dynamics and lifetimes [3].
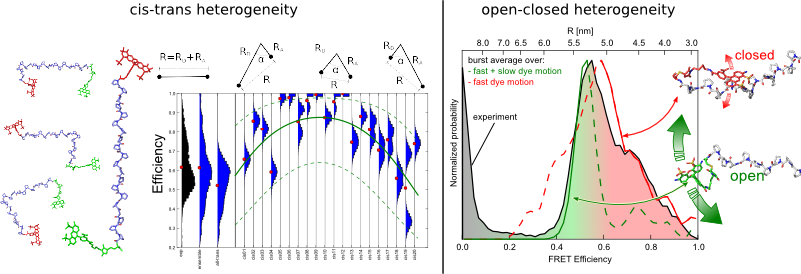
Conformational Heterogeneity
Left: Efficiency histograms of all-trans and cis-isomers. Central cis-bonds reduce the distance and lead to increased FRET efficiency. Right: Efficiency histogram from fast and slow dye motions, corresponding to closed and open conformations of the dyes in comparison with the experiment.
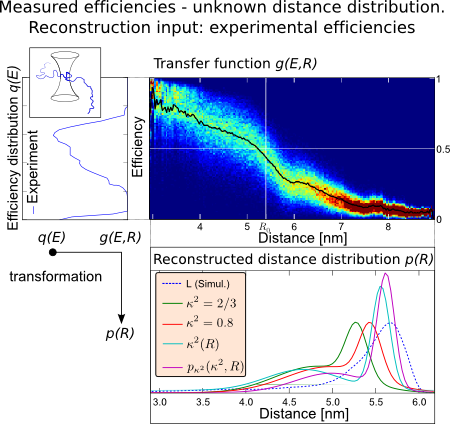
Reconstruction
Distance Reconstruction
Next, we developed a method for distance reconstruction based on transfer functions \(g(E,R)\) \[q(E) = \int\limits_0^{R_{max}} g(E,R) p(R) dR\quad,\]with \(q(E)\) as efficiency and \(p(R)\) as distance distribution. The transfer function formalism describes a convolution of the distance distribution with an arbitrary convolution kernel. In the simplest case, the Förster equation (1) is the convolution kernel. Proceeding from this, we altered the convolution kernel by reducing the approximations in Eq. (1) and replacing them by orientaion information from our simulations[3]. On the right side, an exemplary reconstruction is shown. The Experimental efficiency distribution is transformed by the transfer matrix (rainbow color). The resulting distance distributions for different levels of approximation are shown on the bottom. As seen by comparison to the reference distance distribution, each removal of an approximation systematically improves the accuracy of reconstructed distances.
References:
- Förster, T. Zwischenmolekulare Energiewanderung und Fluoreszens Ann. Phys., 1948, 2, 55-75
- VanBeek, D. B.; Zwier, M. C.; Shorb, J. M. & Krueger, B. P. Fretting about FRET: correlation between kappa and R. Biophys J, 2007, 92, 4168-4178
- Hoefling, M.; Lima, N.; Hänni, D.; Schuler, B.; Seidel, C. A. M. & Grubmüller, H. Structural Heterogeneity and Quantitative FRET Efficiency Distributions of Polyprolines through a Hybrid Atomistic Simulation and Monte Carlo Approach PLoS ONE, 2011, 6, e19791
- Stryer, L. & Haugland, R. P. Energy transfer: a spectroscopic ruler. Proc Natl Acad Sci U S A, 1967, 58, 719-726
- Schuler, B.; Lipman, E. A.; Steinbach, P. J.; Kumke, M. & Eaton, W. A. Polyproline and the “spectroscopic ruler” revisited with single-molecule fluorescence. Proc Natl Acad Sci U S A, 2005, 102, 2754-2759
- Pool, R.; Heringa, J.; Hoefling, M.; Schulz, R.; Smith, J. C. & Feenstra, K. A. Enabling grand-canonical Monte Carlo: Extending the flexibility of GROMACS through the GromPy python interface module. J Comput Chem, 2012